Why Self-Hosted LLMs May Be the Future for Clinical Research
Large Language Models (LLMs) are quickly transforming how we process information across every industry, especially in life sciences, and the capabilities are impressive. LLMs are used in a variety of ways, like summarizing complex data or generating research insights, for instance. However, when working with highly-sensitive data like patient-level clinical trial results, the balance between performance, privacy, and control becomes critical.
In our PHUSE US Connect award-winning paper on the same topic, we explored one important question: Can open-access, self-hosted LLMs realistically compete with large-scale, closed-source models, while still protecting data and maintaining control?
Our research suggests they can, and the results are promising.
Turning Complexity into Clarity with Topological Data Analysis
Topological Data Analysis (TDA) is a relatively new approach for analyzing complex, high-dimensional data. Unlike traditional statistical methods that often rely on assumptions about distribution or linearity, TDA uses geometry and topology to extract meaningful patterns from data.
At its core, TDA transforms raw clinical data into a visual network or graph. Each node in the graph usually represents one patient, and edges connect nodes based on similarity across selected clinical outcomes. This structure allows researchers to identify “communities,” subgroups of patients who share similar characteristics or treatment responses. These communities may reveal insights that would be difficult or impossible to detect using conventional methods.
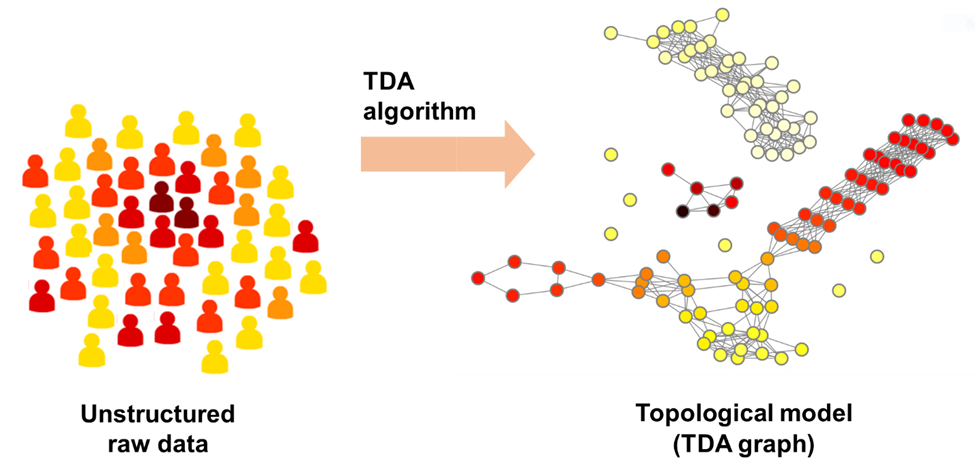
For instance, two patients may appear unrelated when viewed through a single variable like blood pressure. However, when evaluated across multiple outcomes such as lab results, mental health scores, and demographic data, they might share a deeper clinical resemblance. TDA captures these relationships by assessing the proximity of patients in a multidimensional space, then converting those findings into an interpretable visual graph.
Our TDA-based workflow for analyzing clinical data reviews through a series of automatic steps includes data pre-processing, selection of outcomes and predictors, construction of the topological model (a graph), identifying communities (signals), and finally, a robust statistical analysis of the found communities. This results in a highly granular, richly annotated dataset that requires sophisticated summarization and interpretation.
This is where LLMs and AI come into play. By integrating the structured insights from TDA into a Retrieval-Augmented Generation workflow, we aimed to automate the generation of research summaries and comparisons that would otherwise require extensive manual effort and subject-matter expertise.
Read the full paper to see how we did it.
Understanding the Risk of Closed-Source AI
Closed-source LLMs like GPT, Claude, and Gemini have set the bar for performance that many in the general population have come to expect. These models are trained on massive datasets and backed by extensive infrastructure, making them ideal for general-purpose tasks where speed, fluency, and creativity are required.
However, when it comes to regulated industries like ours, these models introduce several critical concerns. The most pressing issue is data privacy. Using closed-source LLMs typically requires sending information to external servers, often through APIs. Even with robust encryption protocols and data handling agreements, this model of interaction places sensitive patient-level information outside the control of the organization.
This is especially problematic in the context of clinical trials, where compliance with regulations like HIPAA, GDPR, and 21 CFR Part 11 must be strictly maintained. Submitting data to a third-party model, no matter how secure, may create unacceptable risk exposure or violate internal policies.
There are also transparency challenges. Users have limited visibility into how the aforementioned models are trained or how responses are generated. This “black box” nature makes it difficult to audit responses or understand potential biases.
Additionally, relying on external providers means giving up a degree of operational control. Updates to the model, changes in pricing, or shifts in access policies are all managed by the vendor. This dependence can create challenges for long-term planning, budgeting, and scalability. While these models excel in many areas, organizations working with sensitive data are right to be cautious. This is why many in the clinical research space are actively exploring alternatives that can be deployed internally and customized to meet their specific needs.
A Smarter Approach with Self-Hosting and Retrieval-Augmented Generation
To preserve data integrity and control, we evaluated open-access models including Llama 3.1 (Meta), Phi-4 (Microsoft), and Gemma 2 (Google). To boost their effectiveness, we implemented a Retrieval-Augmented Generation (RAG) pipeline. This technique enriches model prompts with relevant, pre-processed content drawn from a local vector database.
The pipeline involved several key steps:
- Preprocessing TDA results into structured text
- Embedding those texts into a vector database
- Dynamically retrieving context based on the user prompt
- Feeding that information to the LLM for response generation
- Evaluating output quality through both expert and automated reviews
We tested this system using a dataset from the NIDA-0048 clinical trial from the National Institute of Drug Abuse (NIDA). Questions were designed to assess performance across multiple tasks, including summarization, comparison, hypothesis generation, and pattern recognition.
Closed-source models still led the way, with Claude 3.5 Sonnet achieving the highest scores and Gemini 2.0 and GPT-4o mini getting the lowest scores. However, Phi-4, a locally hosted open-access model, performed impressively when enhanced with the RAG pipeline. It achieved 85 percent of Gemini’s average performance, even outperforming in some task areas like contextual comparison.
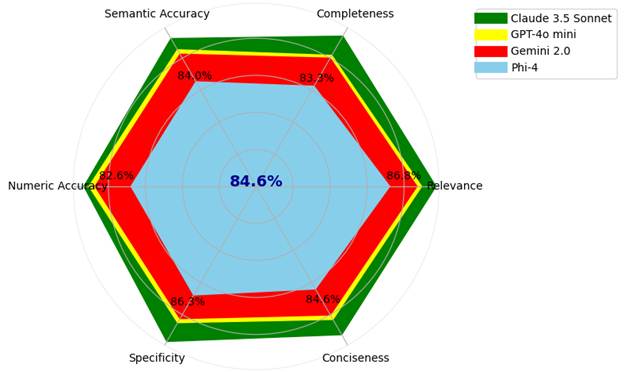
These results demonstrate that open-access models, when properly enhanced and supported by the right infrastructure, can be serious contenders. For clinical organizations that need to keep data in-house, this is a breakthrough.
Better Evaluation Tools for More Reliable Insights
During testing, we found that even top LLMs struggled with numerical accuracy. To improve evaluation, we developed a custom tool called the NumberContextValidator (NCV). This tool checks semantic and numeric correctness using strict criteria, which is essential when working with clinical trial data that demands precision.
Looking ahead, the rapid evolution of LLMs demands a thoughtful approach from the pharmaceutical industry to prioritize data security, scientific accuracy, and operational flexibility. What this study shows is that organizations no longer have to choose between cutting-edge performance and full data control.
With the right infrastructure and enhancements like Retrieval-Augmented Generation, open-access, self-hosted LLMs can rise to deliver reliable insights, support clinical workflows, and reduce manual burdens. This is all possible without compromising privacy or compliance.
The future of AI conversations in our industry should focus on building smarter, more adaptable systems that fit the needs of your data and your mission.Click here to read the full paper for deeper insights, model comparisons, and our complete methodology.